This page provides
an implementation of the "pair
hidden Markov models on tree structures (PHMMTSs)",
which is an extension of the
"pair hidden Markov models
(PHMMs)" defined on alignments of trees.
PHMMTS provides a
unifying framework and an automata-theoretic model for alignments
of trees, structural alignments and pair stochastic context-free
grammars.
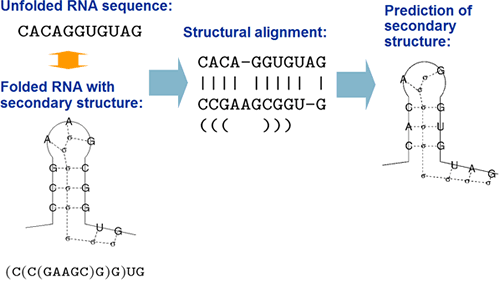
By structural alignment, we mean a pairwise alignment to
align "an unfolded RNA sequence" into "an RNA
sequence of known secondary structure".
|
